Integrative splicing quantitative trait locus analysis reveals risk loci for non-small-cell lung cancer
Recently, Professor Ma Hongxia and Professor Shen Hongbing published an article entitled “Integrative splicing quantitative trait locus analysis reveals risk loci for non-small-cell lung cancer” in The American Journal of Human Genetics. In this study, they first performed splicing quantitative trait locus (sQTL) analysis to systematically investigate the genetic control of alternative splicing by using a repository of genome-wide genotype and gene splicing data in normal lung tissues from 116 donors of Chinese ancestry. Genomic properties of these sQTLs were characterized. Then, they integrated lung sQTLs and the large-scale non-small cell lung cancer (NSCLC) GWAS (13,327 cases and 13,328 controls) by using splice-transcriptome-wide association study (spTWAS)to uncover susceptibility loci of NSCLC. Finally, functional experiments were carried out to confirm the biological mechanisms of the potential causative variant and target gene. (Figure 1).
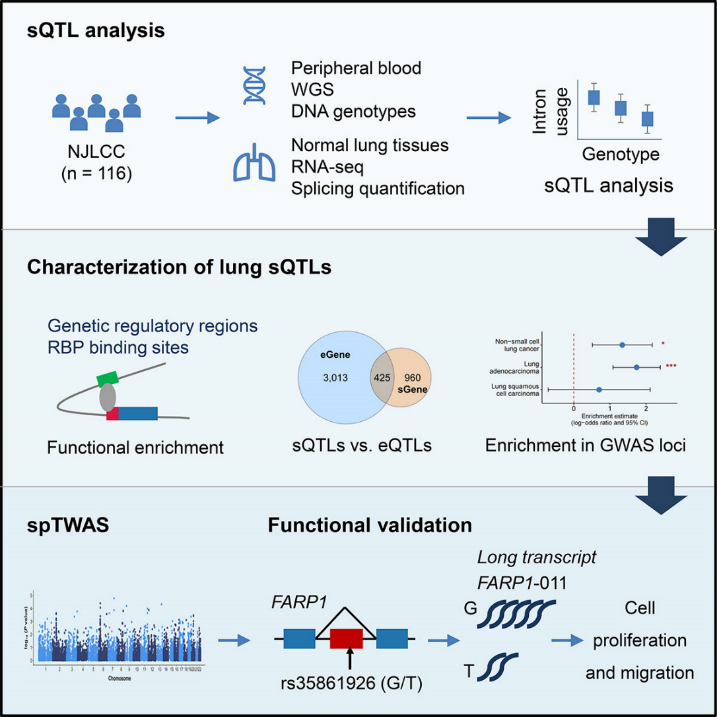
Figure 1. A flow chart of the study design
Lung cancer is one of the most commonly diagnosed cancers and the leading cause of cancer mortality in China. NSCLC accounts for approximately 85% of total lung cancer cases. The development of lung cancer is driven by multiple factors involving environmental exposures and germline genetic variants. Since 2008, genome-wide association studies (GWASs) have identified 61 susceptibility loci for lung cancer, which provided important insights into the genetic architecture of lung cancer. However, GWAS risk variants account for only a modest proportion of the estimated heritability of lung cancer. Furthermore, since the majority of risk variants are located in non-coding regions of the genome, the target genes and downstream biological pathways that mediate these associations remains elusive.Alternative splicing is a crucial post-transcriptional regulatory mechanism, which allows a single pre-mRNA to produce multiple mature mRNA isoforms that can be translated into functionally diverse proteins. More than 95% of human genes are affected by alternative splicing. Furthermore, aberrantsplicing patterns are frequently observed in the development and progress of diseases including lung cancer. Increasing evidence has demonstrated that alternative splicing can be modulated by inheritable genetic variants (splice QTL or sQTL). In particular, the identification of sQTLs could help to gain insight into the mechanisms underlying GWAS associations for a number of traits or diseases.
The authors performed sQTL analysis by using a repository of genome-wide genotype and gene splicing data in normal lung tissues from 116 donors of Chinese ancestry, which identified 1,385 sGenes and 378,210 significant variant-intron pairs that contained 3,232 sIntrons. They comprehensively characterized the genomic features of sQTLs, which found that sQTLs were clustered around splice site, enriched in actively transcribed regions, genetic regulatory elements (e.g., promoters, enhancers and transcription factor binding sites) and splicing factor binding sites. Moreover, sQTLs were largely distinct from expression quantitative trait loci (eQTLs) and showed significant enrichment in potential risk loci of NSCLC. (Figure 2)
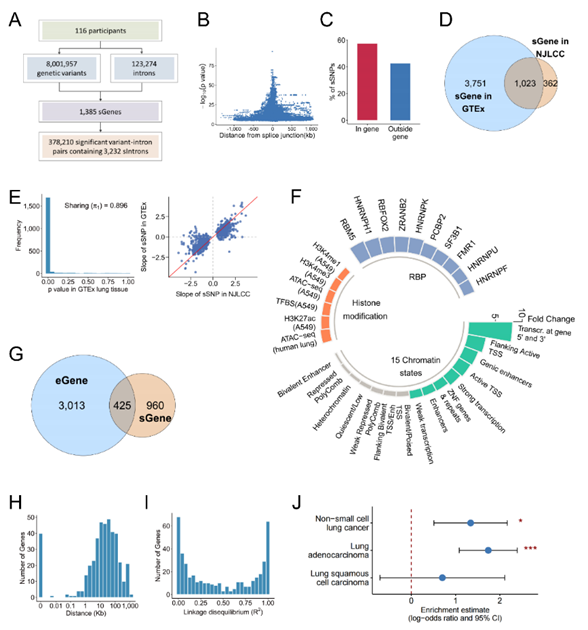
Figure 2. Identification and characterization of sQTLs, comparison of sQTLs and eQTLs, and enrichment of NSCLC GWAS variants in lung sQTLs.
To identify susceptibility genes of NSCLC, the authors performed spTWAS by integrating data of genotypes and intron usage ratios as the reference panel to reanalyze summary level data of NSCLC GWAS.spTWAS identified 23 alternative splicing events in 19 genes that were significantly associated with the risk of overall NSCLC or histological subtypes. Among those loci, 7q22.3 (RP11-325F22.2), 3q23 (XRN1), 8q23.1 (EIF3E), and 13q32.2 (FARP1) were newly identified risk loci. Colocalization analysis showed that two significant alternative splicing events at 8q23.1 and one at 13q32.2 were highly likely to colocalize (PP4 > 0.7).
Alternative splicing events of EIF3E in 8q23.1 were found to be significantly associated with the risk of lung adenocarcinoma. Conditional analyses showed that intron EIF3E:chr8:109,245,901:109,247,227 largely explained the GWAS signal at this region. Furthermore, GWAS signal of lung adenocarcinoma, with lead variant rs443680, colocalized with the sQTL of this splicing event. Bioinformatics analyses indicated that EIF3E splicing, which modified the expression of EIF3E-011 but not the total expression of EIF3E, might be a mediator of the link between genetic variations in 8q23.1 and risk of lung adenocarcinoma. (Figure 3)
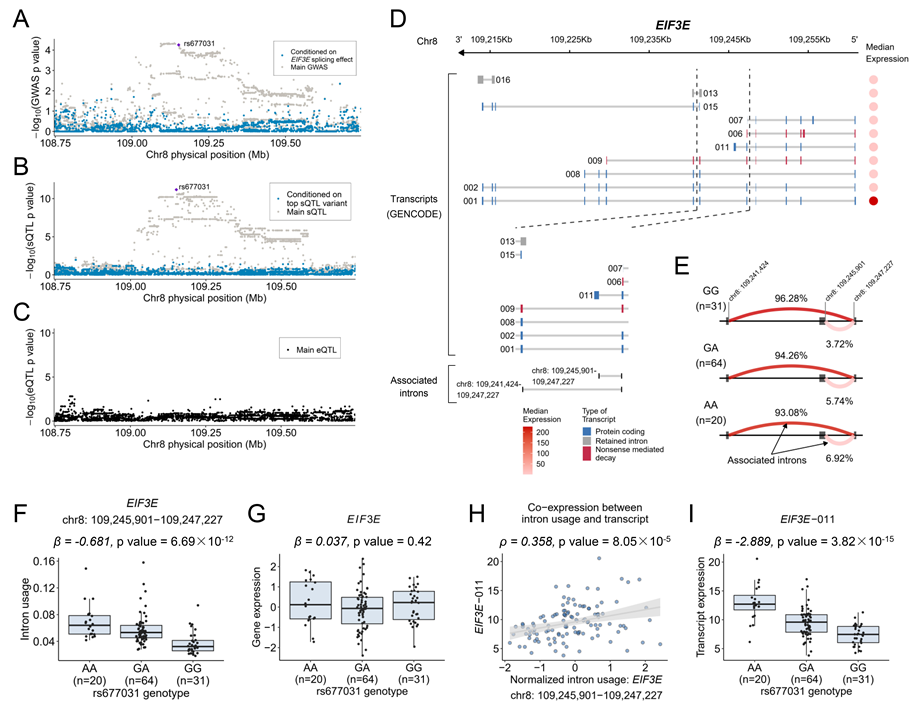
Figure 3. spTWAS associations at EIF3E implicates a target gene independent of genetic effects on total expression.
Alternative splicing event of FARP1 (FARP1:chr13:99,090,112:99,091,058) in 13q32.2 was found to be significantly associated with the risk of lung adenocarcinoma. The most significant variant rs35861926 in 13q32.2 that was associated with risk of lung adenocarcinoma (rs35861926-T, OR = 0.88, 95%CI:0.82-0.93, P = 1.87×10-5) was also the sSNP for intron FARP1:chr13:99,090,112:99,091,058. The rs35861926-T allele was associated with a decreased usage of intron FARP1:chr13:99,090,112:99,091,058 and a decreased expression of FARP1-011 in normal lung tissues. Further functional annotation and experiments confirmed that rs35861926-T can reduce the risk of lung adenocarcinoma by promoting FARP1 exon 20 skipping to down-regulate the expression level of the long transcript FARP1-011. Overexpression of FARP1-011 promoted the migration and proliferation of lung adenocarcinoma cells.
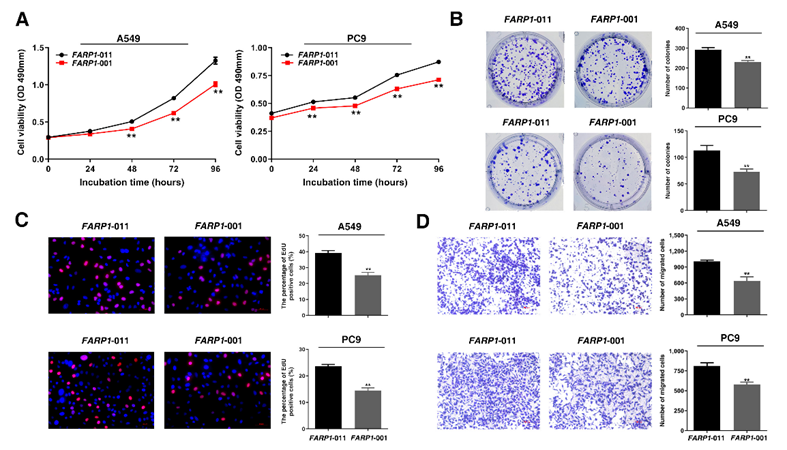
Figure 4. spTWAS association at FARP1 implicates a target gene independent of genetic effects on total expression.
Overall, our study provided a comprehensive catalog of lung sQTLs, which provided an informative lung sQTL database for investigating the genetic mechanisms of lung diseases. In addition, a combination of spTWAS analyses and functional experiments identified novel risk loci of lung adenocarcinoma and provided additional insights into the molecular mechanisms underlying the risk loci.
Journal: American Journal of Human Genetics
Title: Integrative splicing quantitative trait locus analysis reveals risk loci for non-small-cell lung cancer
Link: https://www.cell.com/ajhg/fulltext/S0002-9297(23)00248-3?journal=ajhg&publicationCode=ajhg&jc=ajhg

