Integration of Non-coding RNA and DNA Methylation in Pan-cancer
Recently, Professor Chen Feng's research group at the School of Public Health, Nanjing Medical University, published a research paper titled "Large- scale integration of the noncoding RNAs with
DNA methylation in human cancers" in the journal CellReports.
Associate Professor Shen Sipeng from the Department of Biostatistics at the School of Public Health served as the first and corresponding author, doctoral student Chen Jiajin as co-first author, Professor Zhao Yang as the main corresponding author, and Professor Chen Feng as a senior author.

Cancer is a complex disease, and DNA methylation, a form of epigenetic modification, regulates gene expression, genome stability, and cell activity. Non- coding RNAs (ncRNAs), including
miRNAs, lncRNAs, rRNAs, snRNAs, and snoRNAs, function in biological regulation at the RNA level. Characterizing the effects of ncRNA quantitative trait methylation sites (ncQTM) and
evaluating their causal roles in cancer and other biological processes hold significant public health and clinical value.
The research group categorized ncQTM into two modules for investigation: cis (ncRNAs from DNA methylation sites within ≤1Mb) and trans (ncRNAs from DNA methylation sites >1Mb or
interchromosomal). Analysis was conducted on 8,545 patients across 32 cancer types from The Cancer Genome Atlas (TCGA) database. Additionally, comparison was made between 763 tumor
tissues from the Clinical Proteomic Tumor Analysis Consortium (CPTAC) and 516 normal tissues from the Genotype-Tissue Expression (GTEx) database.

Figure1. Research Workflow
Through over 22 billion association analyses, 302,764 cis- ncQTM and 79,841,728 trans- ncQTM were ultimately identified. Among the cis results, the most associated non-coding RNAs included LINC00982, PSMB8-AS1,HOXA11-AS, HOXC-AS3, and HOTAIR. In the trans results, the most associated non-coding RNAs included PCED1B-AS1, LINC00355, RP11-356I2.4, DSCR4-IT1, and ST8SIA6-AS1. Furthermore, the majority of trans associations (88.9%) were interchromosomal.


Figure2. Association results of cis and trans ncQTM
In order to efficiently utilize the comprehensive resources of ncQTM across 32 human cancertypes, the research group has developed a Pancan-ncQTM database:http://bigdata.njmu.edu.cn/Pancan-ncQTM/.
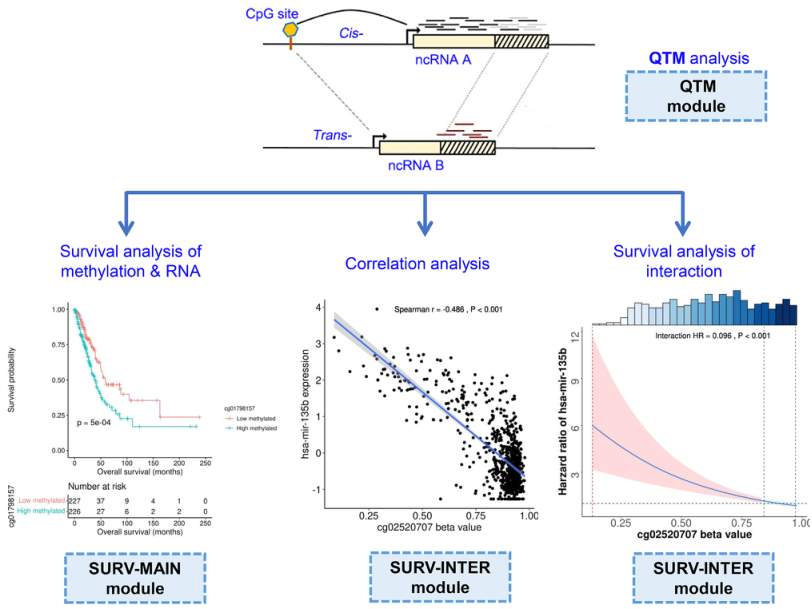
Figure3. Pancan-ncQTM Database
Date: March 2023
Journal: Cell Reports
Title:Large-scale integration of the non-coding RNAs with DNAmethylation in human cancers Original Article:https://doi.org/10.1016/j.celrep.2023.112261

